Viruses are ubiquitous: for every organism, there are multiple viruses that can infect it. Although we know much about the beautiful structures of viruses, and we understand how certain viruses (the ones that are bad for us) infect a cell, we know little about the dynamics of these organisms. We want to understand the life cycle of viruses from a physical point of view. We are particularly interested in understanding how they self-assemble.
The video below (part of a press release from Harvard SEAS) explains some of our recent work on developing optical tools to see viruses:
The simplest viruses consist of only of a single molecule of genomic nucleic acid (DNA or RNA) surrounded by a highly-symmetric protective shell composed of many copies of a single protein (the capsid protein). The structures of mature viruses are known in exquisite, atomic-resolution detail, but the mechanisms by which they form remain a mystery. Many viruses spontaneously assemble themselves within the interior of a host cell by self-assembly, a process that is fundamentally random. How do these perfectly-formed, infectious viruses emerge so efficiently from a randomly fluctuating soup of capsid protein and viral genome? And, once formed, how do they release of their genomes in order to initiate an infection?
We are addressing these questions by developing optical techniques that allow us to follow individual virus particles, their proteins, and their genomes, in three dimensions and in real time. We use variations on holographic microscopy and elastic light scattering to do fast, sensitive measurements of single viruses as they assemble and disassemble in vitro.
Elastic scattering is fast, in principle. One can resolve the dynamics to sub-millisecond precision over long periods of time. In practice, however, the background signal from reflected light, dust, or imperfections in the optical path can easily overwhelm the weak scattering from the viruses. Together with collaborators from the Leiden University, Heraeus Quarzglas, the Leibniz Intitute of Photonic Technology, and the Otto Schott Institute of Material Research, we developed a new technique to overcome these experimental limitations, which we call the "fiberscope".
Using the fiberscope, we have been able to track individual unlabeled viruses 26 nm in diameter at rates of thousands of measurements per second (see below). These are the smallest viruses to be tracked on sub-millisecond time scales, which are comparable to the time scales for self-assembly. We are currently working on improving our technique to detect single capsid proteins and follow their assembly.
For more details, see our paper in ACS Nano.
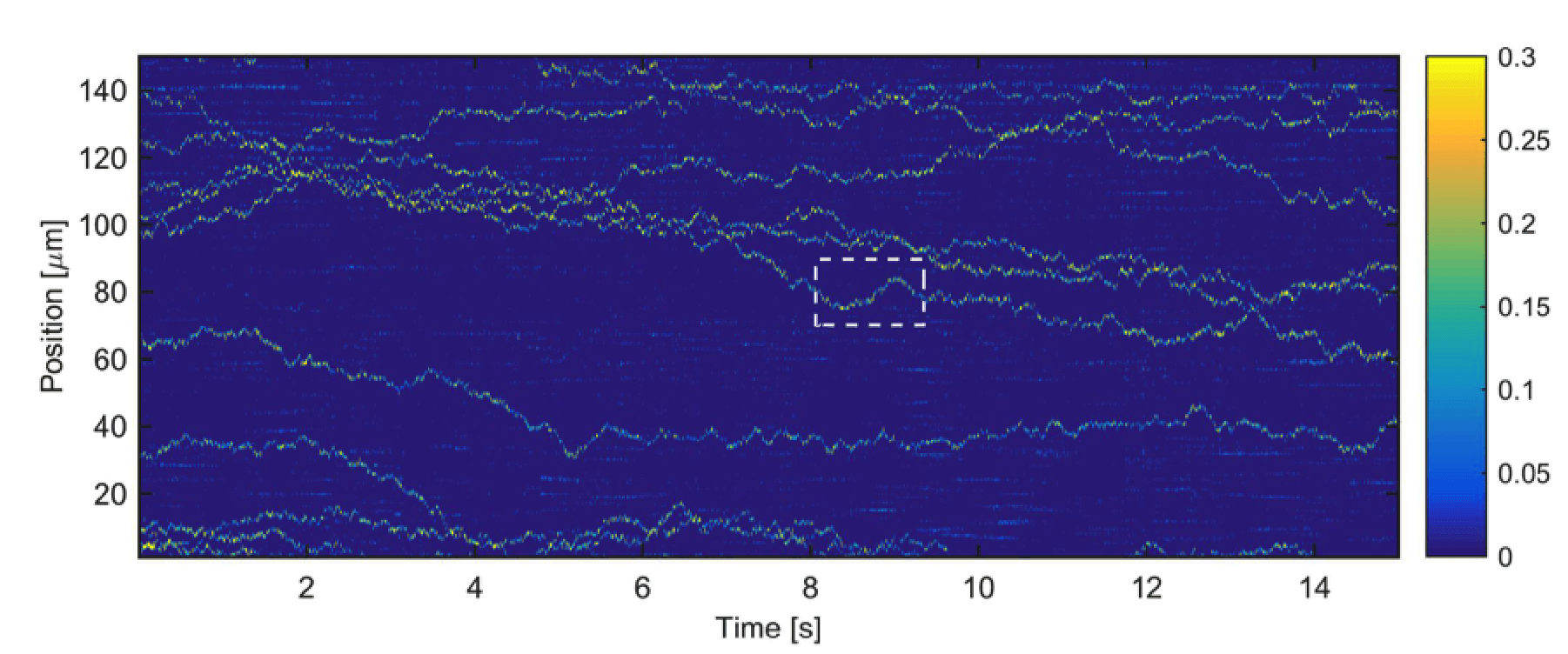 Tracking single, unlabeled, freely diffusing CCMV virions. The color bar indicates the signal level compared to the background. Measurements are taken at 2 KHz, a rate comparable to the timescales of self-assembly.
Tracking single, unlabeled, freely diffusing CCMV virions. The color bar indicates the signal level compared to the background. Measurements are taken at 2 KHz, a rate comparable to the timescales of self-assembly.


